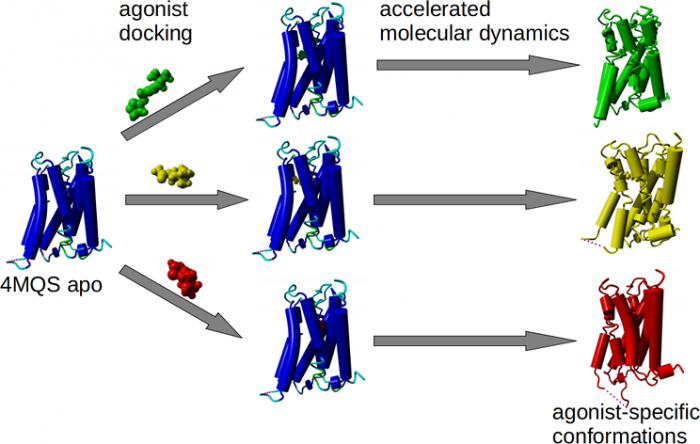
Agonist-specific conformations of the M<sub>2</sub> muscarinic acetylcholine receptor assessed by molecular dynamics

Structurally diverse agonists for a given receptor induce receptor conformations specific to each structural family. These agonist-specific conformations can lead to non-uniform modulation of signalling pathways. This preferential orientation of signalling of a given receptor towards a subset of its signal transducers is termed signalling bias. This property may be employed to develop drugs that selectively produce desired effects while avoiding side effects associated with activation of unwanted signalling pathways. We modelled conformations of the M2 receptor specific to individual agonists, including the newly developed Gi-biased agonists.
Binding of muscarinic ligands, both antagonists and agonists, and their effects on the conformation of the M2 acetylcholine receptor were modelled in silico and compared to experimental data. After docking of antagonists to the M2 receptor in an inactive conformation (3UON, 5ZK3, 5ZKB or 5ZKB) and agonists in an active conformation (4MQS) 100 ns of conventional molecular dynamics (MD) followed by 500 ns of accelerated MD was run. Conventional MD revealed ligand-specific interactions with the receptor. Antagonists stabilized the receptor in an inactive conformation during accelerated MD. The receptor in complex with various agonists attained different conformations specific to individual agonists. The magnitude of TM6 movement correlated with agonist efficacy at non-preferential Gs pathway. The shape of the intracellular opening where the receptor interacts with a G-protein was different for the classical agonist carbachol, super-agonist iperoxo and Gi/o-biased partial agonists JR-6 and JR-7, being compatible with experimentally observed agonist bias at the G-protein level. Moreover, wash-resistant binding of the unique agonist xanomeline associated with interaction with membrane lipids was formed during accelerated MD. Thus, accelerated MD is suitable for modelling of ligand-specific receptor binding and receptor conformations that is essential for the design of experiments aimed at the identification of the secondary binding sites and understanding molecular mechanisms underlying receptor activation.
