Laboratory of Structural Biology of Signaling Proteins
Content of this page

About the Laboratory
Our Laboratory is focused on structural biology (the relationship between the structure and function of certain groups of proteins), particularly we focus on the proteins which participate in the signal transmission in the cell. Among methods we use are recombinant protein expression, biophysical characterization, study of intermolecular interactions, protein structure and interaction surfaces. All these methods enable us to better understand the details how the activity and function of protein-protein complexes is regulated. Our research is focused mainly on:
- Structural biology of 14-3-3 proteins and their complexes
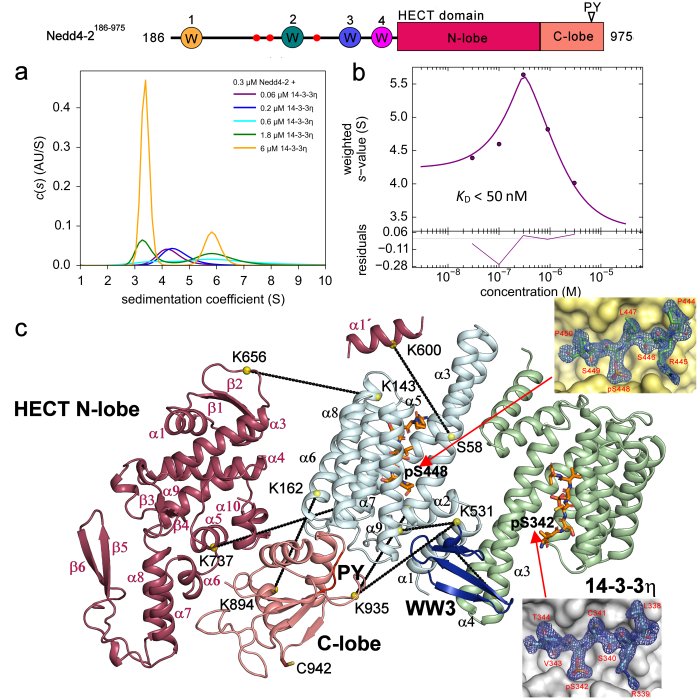
- Study of inhibition of ubiquitin ligase Nedd4-2 by 14-3-3 protein
- Characterizaton of the interactions between transcription factor FOXO4 and tumour supressor p53
- Mechanism of regulation of proteinkinase ASK1 by TRX and 14-3-3 protein
- Mechanism of regulation of protease caspase-2 by 14-3-3 protein
Projects
Achievements
Otto Wichterle Prize
Prize of Otto Wichterle for Dr. Dalibor Košek.
Dalibor Kosek received the prize of Otto Wichterle for excellent young scientists of CAS in the year 2022.
NEW FINDINGS ON THE STRUCTURE OF THE FOXO4: p53 COMPLEX - A KEY FACTOR IN SENESCENCE REGULATION (9.4.2022)
Transcription factor p53 protects cells against tumorigenesis when subjected to various cellular stresses. Under stress conditions, p53 interacts with another transcription factor, FOXO4 (Forkhead box O 4), and together they increase the production of p21 protein, which triggers the process of cell aging (senescence). However, the molecular mechanism of upregulation of p21 transcription is still unclear. In a work published in the Protein Science journal, scientific teams of Dr. Obsilova (IPHYS CAS), prof. Obsil (Faculty of Science, Charles University and IPHYS CAS) and their colleagues from IOCB CAS characterized interactions between p53 and FOXO4 at the molecular level. New knowledge about the structure of the complex may enable the development of specific inhibitors of the interaction between these two proteins, and subsequently in the development of new drugs aimed at the selective elimination of senescent cells.
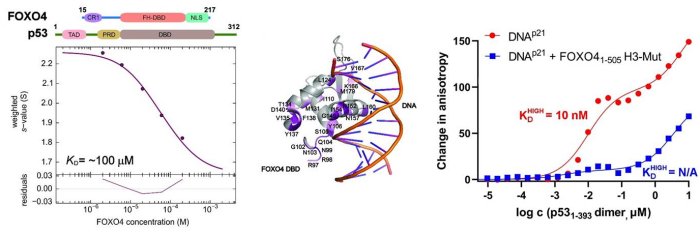
In this structural study, the researchers performed a detailed characterization of the interactions in the FOXO4: p53 complex using an integrated approach involving analytical ultracentrifugation, nuclear magnetic resonance, and chemical cross-linking coupled to mass spectrometry. Because both FOXO4 and p53 have multiple domains (see Figure), they studied the role of individual domains and disordered segments of both proteins and mapped their interaction interfaces. They found out that the interaction between p53 transactivation domain TAD and the FOXO4 Forkhead domain is crucial for the overall stability of the p53:FOXO4 complex. Furthermore, contacts involving the N-terminal disordered FOXO4 segment, the C-terminal negative regulatory domain of p53, and the DNA-binding domains of both proteins stabilize the complex formation. By measuring DNA binding, they further found that the p53: FOXO4 complex formation blocks p53 binding to DNA without affecting the DNA-binding properties of FOXO4.
Left, sedimentation velocity analytical ultracentrifugation analysis of interaction between FOXO4 and p53. Middle, chemical shift perturbations obtained from 1H-15N HSQC spectra of 15N-labeled FOXO4 in the presence of p53 mapped onto the crystal structure of the FOXO4 DBD:DNA complex. Right, fluorescence anisotropy measurements showing that the complex formation reduces the DNA-binding affinity of p53.
Mandal R, Kohoutova K, Petrvalska O, Horvath M, Srb P, Veverka V, Obsilova V and Obsil T. FOXO4 interacts with p53 TAD and CRD and inhibits its binding to DNA. Protein Sci. roč. 31, č. 5 (2022), č. článku e4287. IF = 6.725. DOI: 10.1002/pro.4287.
Our new articles in Communications Biology

Two accepted papers in Communications Biology: July and August 2021
Pavel Pohl, Rohit Joshi, Olivia Petrvalska, Tomas Obsil and Veronika Obsilova
Commun. Biol. 2021 July 22; 4(1):899.
Matej Horvath, Olivia Petrvalska, Petr Herman, Tomas Obsil and Veronika Obsilova
Commun. Biol. 2021 August 19; 4(1):986.
IF = 6.268
Innovative method of the Forhkead transcription factor FOXO3 regulation
The study published in prestigious journal eLife identifies small molecule compounds that interact with the Forkhead box O3 transcription factor (FOXO3) and modulate its activity.
FOXO3, with the characteristic fork head DNA-binding domain is part of the O subclass of the forkhead family of transcription factors. These transcription factors have important roles in mammalian cells in regards to regulating cell homeostasis, differentiation, longevity and steer cell death. The activity of FOXO3 in particular contributes to therapy-resistance programs that protect cancer cells during chemo and radiotherapy. Recent studies have also found the DNA-binding domain (DBD) of FOXO aid protein-protein interactions with other key regulators of longevity and death, and drug resistance. A reversible inhibition of FOXO3 activity by small compounds thereby might boost anti-tumor immune responses.
In order to inhibit the FOXO3 activity, it was first necessary to identify small molecule compounds that could block the interaction between FOXO3 and DNA. Using the structural data of FOXO3 DBD and FOXO4 DBD, the researchers developed six different pharmacophore models that were used for in silico screening of small molecule compound databases. Selected compounds were then tested for their ability to inhibit FOXO3 function both in vitro, and in cancer cell lines. The interactions of these compounds with FOXO3 DBD were assessed using NMR spectroscopy and docking studies.
The teams of Dr. V. Obšilová from the Institute of Physiology CAS in BIOCEV and prof. T. Obšil from the Faculty of Science, Charles University with the team of prof. M. J. Ausserlechner from the Medical University of Innsbruck, Austria identified the compounds S9 and its oxalate salt S9OX as compounds able to inhibit the FOXO3 activity in cancer cells. They also proposed that due to their mode of binding to FOXO3 DBD, these compounds may also interfere with protein-protein interactions of FOXO3. The advantage of these compounds is the strict control of application-dose and -time and the fact that they are not immunogenic allowing repeated applications – so dose- and application time can be adjusted to damage cancer cells or boost anti-cancer immunity, but also limit unwanted side effects of FOXO-inhibition on stem cells and other somatic tissues. Future research will be focused on investigation whether or not S9 can be used as a chemical foundation for developing FOXO-regulatory compounds that would control the functions and target gene subsets of FOXO transcription factors.
Original paper: Hagenbuchner J*, Obsilova V*, Kaserer T*, Kaiser N, Rass B, Psenakova K, Docekal V, Alblova M, Kohoutova K, Schuster D, Aneichyk T, Vesely J, Obexer P, Obsil T+, Ausserlechner MJ+. Modulating FOXO3 transcriptional activity by small, DBD-binding molecules. eLife. 2019, 8(Dec 4), e48876. doi: 10.7554/eLife.48876. IF = 7.551
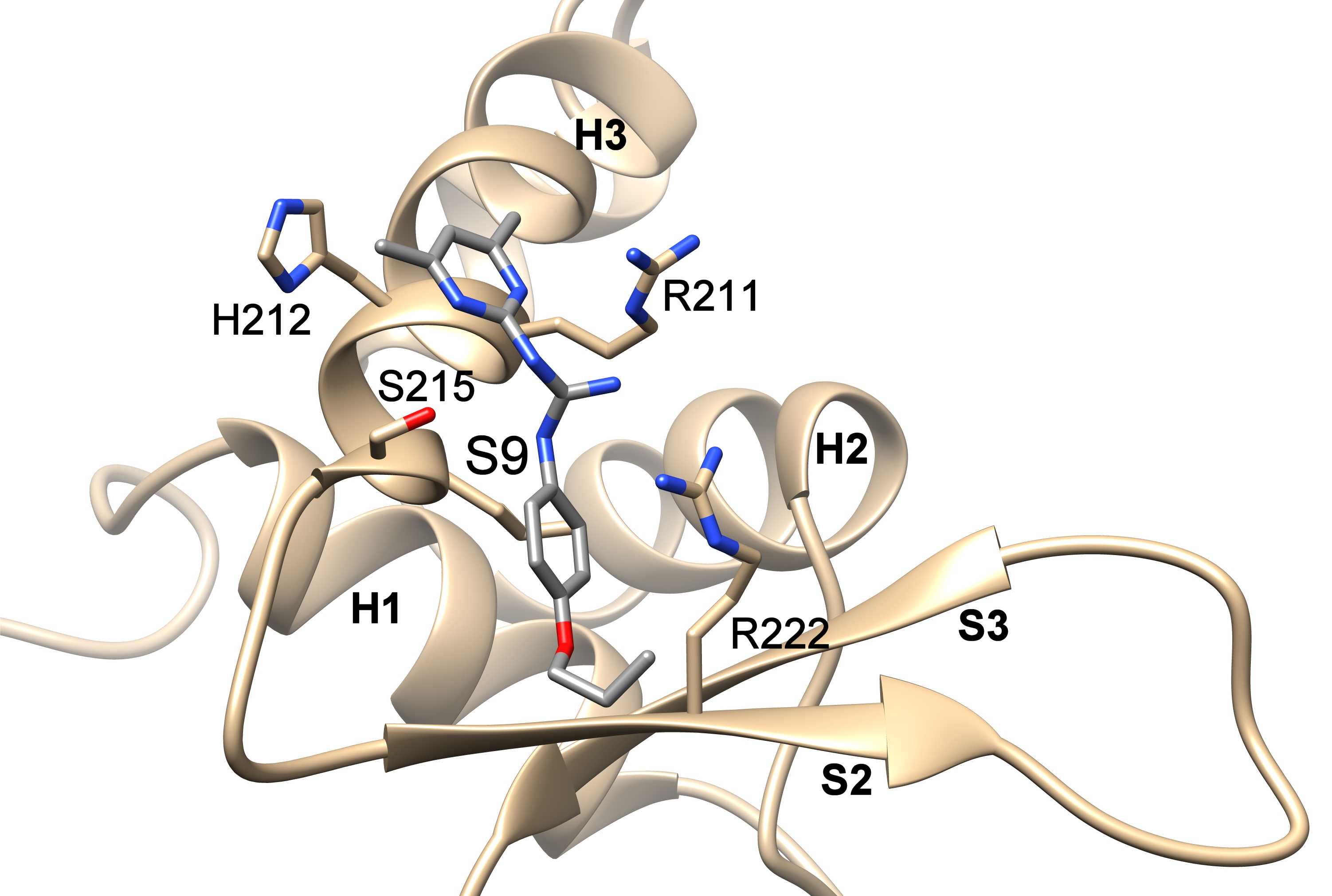
Compounds S9 blocks the DNA binding surface of Forkhead transcription factor FOXO3. The figure shows the structural model of the DNA-binding domain of FOXO3 with bound compound S9 based on data from NMR measurements and docking simulations.
The best publication in Physiological Research with the young authors for the year 2018
We obtained the first prize for the best publication in Physiological Research with the young authors from for the year 2018.
The best publication with the authors from IP for the year 2017
We got the prize for the best publication of the Institute of Physiology for year 2017 for the publication:
Alblova M, Smidova A, Docekal V, Vesely J, Herman P, Obsilova V, Obsil T. :
Molecular basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1.
Proc Natl Acad Sci U S A. (2017) 114:E9811-E9820. IF = 9.504.
Our new publication in Journal of Biological Chemistry: Role of the EF-hand-like Motif in the 14-3-3 Protein-mediated Activation of Yeast Neutral Trehalase Nth1.
Kopecka M, Kosek D, Kukacka Z, Rezabkova L, Man P, Novak P, Obsil T, Obsilova V.
J Biol Chem. 2014 May 16;289(20):13948-61.
http://www.ncbi.nlm.nih.gov/pubmed/24713696
Trehalases hydrolyze the non-reducing disaccharide trehalose amassed by cells as a universal protectant and storage carbohydrate. Recently, it has been shown that the activity of neutral trehalase Nth1 from Saccharomyces cerevisiae is mediated by the 14-3-3 protein binding that modulates the structure of both the catalytic domain and the region containing the EF-hand-like motif, whose role in the activation of Nth1 is unclear. In this work, the structure of the Nth1·14-3-3 complex and the importance of the EF-hand-like motif were investigated using site-directed mutagenesis, hydrogen/deuterium exchange coupled to mass spectrometry, chemical cross-linking, and small angle x-ray scattering. The low resolution structural views of Nth1 alone and the Nth1·14-3-3 complex show that the 14-3-3 protein binding induces a significant structural rearrangement of the whole Nth1 molecule. The EF-hand-like motif-containing region forms a separate domain that interacts with both the 14-3-3 protein and the catalytic trehalase domain. The structural integrity of the EF-hand like motif is essential for the 14-3-3 protein-mediated activation of Nth1, and calcium binding, although not required for the activation, facilitates this process by affecting its structure. Our data suggest that the EF-hand like motif-containing domain functions as the intermediary through which the 14-3-3 protein modulates the function of the catalytic domain of Nth1.
Our New Paper in JBC: Structural Characterization of Phosducin and its Complex with the 14-3-3 Protein
Kacirova M, Kosek D, Kadek A, Man P, Vecer J, Herman P, Obsilova V and Obsil T
J. Biol. Chem. jbc.M115.636563. First Published on May 13, 2015, doi:10.1074/jbc.M115.636563
Kacirova M, Kosek D, Kadek A, Man P, Vecer J, Herman P, Obsilova V and Obsil T
http://www.ncbi.nlm.nih.gov/pubmed/25971962
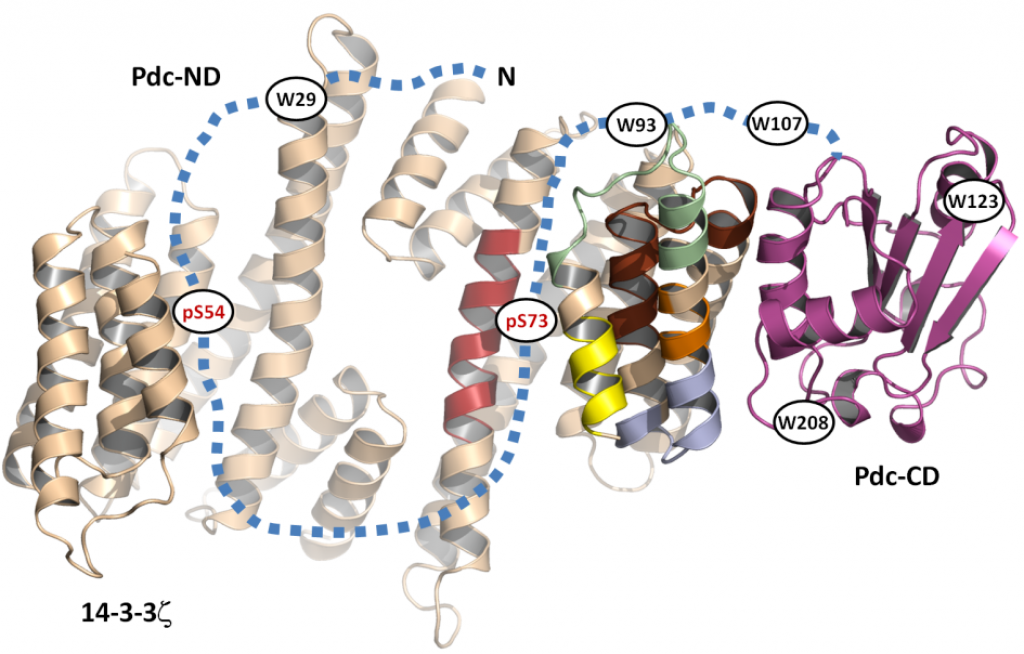
Phosducin (Pdc), a highly conserved phosphoprotein involved in the regulation of retinal phototransduction cascade, transcriptional control, and modulation of blood pressure, is controlled in the phosphorylation-dependent manner including the binding to the 14-3-3 protein. However, the molecular mechanism of this regulation is largely unknown. Here, the solution structure of Pdc and its interaction with the 14-3-3 protein were investigated using small angle X-ray scattering, time-resolved fluorescence spectroscopy and hydrogen-deuterium exchange coupled to mass spectrometry. The 14-3-3 protein dimer interacts with Pdc using surfaces both inside and outside its central channel. The N-terminal domain of Pdc, where both phosphorylation sites and the 14-3-3 binding motifs are located, is intrinsically disordered protein which reduces its flexibility in several regions without undergoing dramatic disorder-to-order transition upon binding to 14-3-3. Our data also indicate that the C-terminal domain of Pdc interacts with the outside surface of the 14-3-3 dimer through region involved in Gtβγ binding. In conclusion, we show that the 14-3-3 protein interacts with and sterically occludes both the N- and C-terminal Gtβγ binding interfaces of phosphorylated Pdc, thus providing a mechanistic explanation for the 14-3-3-dependent inhibition of Pdc function.
Tomáš Obšil was awarded the title of professor
Prof. RNDr. Tomáš Obšil, Ph.D. was awarded by the title of professor at The Faculty of Science, Charles University, majoring in Physical Chemistry. Professors graduation will take place December 18, 2014.
Josef Hlávka Award and Medal for 2014
16. 12. 2014: The Foundation of Josef, Marie and Zdeňek Hlávka awarded Hana Janoušková for being the best graduated student from 2nd Medical Faculty, Charles University in Prague.
Our new paper in JBC: Biophysical and Structural Characterization of the Thioredoxin-binding Domain of Protein Kinase ASK1 and Its Interaction with Reduced Thioredoxin.
Kosek D, Kylarova S, Psenakova K, Rezabkova L, Herman P, Vecer J, Obsilova V, Obsil T.
J Biol Chem. 2014 Aug 29;289(35):24463-74.
http://www.ncbi.nlm.nih.gov/pubmed/25037217
Apoptosis signal-regulating kinase 1 (ASK1), a mitogen-activated protein kinase kinase kinase, plays a key role in the pathogenesis of multiple diseases. Its activity is regulated by thioredoxin (TRX1) but the precise mechanism of this regulation is unclear due to the lack of structural data. Here, we performed biophysical and structural characterization of the TRX1-binding domain of ASK1 (ASK1-TBD) and its complex with reduced TRX1. ASK1-TBD is a monomeric and rigid domain that forms a stable complex with reduced TRX1 with 1:1 molar stoichiometry. The binding interaction does not involve the formation of intermolecular disulfide bonds. Residues from the catalytic WCGPC motif of TRX1 are essential for complex stability with Trp(31) being directly involved in the binding interaction as suggested by time-resolved fluorescence. Small-angle x-ray scattering data reveal a compact and slightly asymmetric shape of ASK1-TBD and suggest reduced TRX1 interacts with this domain through the large binding interface without inducing any dramatic conformational change.
Získání grantu GA ČR
14-10061S – Mechanismus regulace kinasové aktivity proteinkinasy ASK1 (řešitelé: T. Obšil, PřF UK, a V. Obšilová, FgÚ AV ČR, v.v.i.)
Studentský Velemlok pro nejlepšího pedagoga
21. 2. 2014 – doc. RNDr. Tomáš Obšil oceněn studenty PřF UK jako nejlepší vyučující v oboru chemie
Cenu Studentský Velemlok pro nejlepšího pedagoga v roce 2013 převzal docent Tomáš Obšil z rukou děkana Přírodovědecké fakulty Univerzity Karlovy v Praze 21. 2. 2014. Pedagogové jsou na toto ocenění navrhováni studenty na základě výsledků hodnocení jejich výuky ve studentské anketě.
Publications
Kohoutová; Klára - Dočekal; V. - Ausserlechner; M. J. - Kaiser; N. - Tekel; A. - Mandal; R. - Horváth; M. - Obšilová; Veronika - Veselý; J. - Hagenbuchner; J. - Obšil; Tomáš Lengthening the Guanidine–Aryl Linker of Phenylpyrimidinylguanidines Increases Their Potency as Inhibitors of FOXO3-Induced Gene Transcription. ACS Omega. 2022; 7(38); 34632-34646.
IF = 4.2